pISSN : 3058-423X eISSN: 3058-4302
Open Access, Peer-reviewed
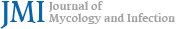
pISSN : 3058-423X eISSN: 3058-4302
Open Access, Peer-reviewed
Waewta Kuwatjanakul,Tran Tuan Khoi,Lumyai Wonglakorn,Kittipan Samerpitak
10.17966/JMI.2024.29.3.137 Epub 2024 October 11
Abstract
Background: The use of a morphological method to determine Aspergillus species in routine laboratory investigation has always been challenged with low sensitivity and low specificity. Time-consuming specimen preparation and insufficient expertise are the crucial reasons. DNA barcoding markers have been introduced for identification as another gold standard method.
Objective: To investigate DNA barcoding markers in identifying Aspergillus strains isolated from patients with eye infection.
Methods: This study extracted 12 DNA samples of 12 Aspergillus strains. DNA barcoding genes, i.e., ITS, beta-tubulin (BT2), calmodulin (CAL), and actin (ACT) genes, were amplified and sequenced by polymerase chain reaction and Sanger's sequencing methods. All gene sequences were compared to those of the type or reference species collected from public databases. The species were identified with phylogenetic analyses in free applications from public websites.
Results: ITS, BT2 and CAL, and ACT identified 5, 6, and 3 species and 3, 2, and 1 species complex of Aspergillus, respectively. DNA barcoding with multi-locus sequence typing revealed four new pathogens of human eye infection, including A. aureoterreus, A. pseudonomiae, A. siamensis, and A. flocculosus-ochraceopetaliformis.
Conclusion: The DNA barcoding markers identified Aspergillus species with better specificity than the conventional method. The markers and techniques need to be used as tools for routine Aspergillus identification.
Keywords
Aspergillus infection DNA barcoding Fungal keratitis New fungal pathogen
The morphological identification of mold in a routine laboratory investigation always struggles with problems of cultivation, specimen preparation, time consumption, and expertise insufficiency in mycological taxonomy. Molecular techniques, such as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and DNA identification, have been introduced to resolve those problems. The MALDI-TOF MS appears more appropriate for routine work because of its faster and simpler techniques but its precision and accuracy remain involved in many factors, i.e., specimen preparation, extraction method, library database, etc. Identification with DNA barcoding markers has been another "gold standard method" for fungal identification1,2. The method includes polymerase chain reaction (PCR), DNA sequencing, and phylogenetic analyses, which have presently been normalized to perform with a lower cost, less complicated techniques, free software, and an available public database. Consequently, the method should be introduced for routine laboratory use.
Fungal keratitis (FK) and endophthalmitis are serious eye infections characterized by a severe sight-threatening con- dition. Over 95% of all FK cases are caused by filamentous fungi. Filamentous fungi have caused >50% of all microbial keratitis with culture-positive cases3,4, and the incidence has increased globally5-8. Over 70 filamentous fungal species have been reported as pathogens. Fusarium spp. and Aspergillus spp. are the most predominant causative agents9,10. The patients with Aspergillus keratitis demonstrated poorer pro-gnosis than those infected by Fusarium and other filamentous fungi11. Aspergillus flavus has been a dominant species to cause Aspergillus keratitis, followed by A. terreus, A. fumigatus, A. niger, A. nidulans, A. ochraceous12-14, which are all prevalent species in human environments. However, some unfamiliar species have caused keratitis such as A. aculeatus15, A. nomiae16, A. viridinutans17, and A. minisclerotigenes18. Many Aspergillus species that thrive in human environments were reported infections in humans. The possibility of inducing eye infection by a new pathogenic species needs to be considered. The present study investigated the potentiality of DNA barcoding markers for identifying 12 Aspergillus strains isolated from patients with eye infections.
1. Collection of Aspergillus strains, morpho- logical identification, and growth observation
A total of 12 Aspergillus strains were isolated from cornea scraping specimens of 12 patients mostly diagnosed with FK. Those patients were admitted to the Srinagarind Hospital, Khon Kaen, Thailand, from January 1, 2022, to December 31, 2022. All Aspergillus strains were presumptively determined into species by morphological characteristics. The optimal growths of all strains were examined by inoculating on blood agar (BA) and Sabouraud dextrose agar (SDA) and incubated at 25℃ and 37℃. The growth of each strain was assessed by measuring a colony-diameter. The data were processed and analyzed with MS Excel 2020 and Statistical Package for the Social Sciences version 20.0 software. The student t-test for continuous variables was conducted to analyze the different radial growth rates between culture media and incubation temperature. A p-value of <0.05 in- dicated a statistical significance.
2. DNA extraction, PCR, and Sanger sequen- cing
A DNA extraction kit (Vivantis Technologies Sdn Bhd., Selangor Darul Ehsan, MY) was used to DNA-extract all strains. NanoDrop 2000 spectrophotometer (Thermo Scien- tific, Massachusetts, USA) was used to quantify DNAs. The internal transcribed spacer (ITS) of the ribosomal gene and partial sequences of actin (ACT), beta-tubulin (BT2), and calmodulin (CAL) genes were amplified by PCR with the primers18-23 shown in Table 1. The PCR reaction contained 10 μM in 1.5 μl for each primer, 12.5 μl of KOD OneTM PCR Master Mix (TOYOBO, JP), and >10 ng of genomic fungal DNA, and added with sterile water to reach a 25 μl of total volume reaction. The PCR condition started at 95℃ for 5 min and was followed by 35 cycles of denaturation at 95℃ for 35 s, annealing at 48℃ for 30 s, extension at 72℃ for 30 s, and amplification in Veriti® Thermal Cycler (Applied Biosystems, CA, USA). The amplification products were checked with 2% gel electrophoresis and submitted to Sanger's sequencing. The Supplementary data file shows the sequences.
|
Genes |
Primers' name |
Primer
pair sequences (5' to 3') |
Ref. |
|
ITS |
ITS1
(Fw) |
TCCGTAGGTGAACCTGCGG |
19 |
|
|
ITS4
(Rw) |
TCCTCCGCTTATTGATATGC |
20 |
|
|
ITS5
(Fw) |
GGAAGTAAAAGTCGTAACAAGG |
20 |
|
|
V9G
(Fw) |
TTACGTCCCTGCCCTTTGTA |
19 |
|
|
LS266
(Rw) |
GCATTCCCAAACAACTCGACTC |
21 |
|
Actin |
ACT-512F
(Fw) |
ATGTGCAAGGCCGGTTTCGC |
22 |
|
(ACT) |
ACT-783R
(Rw) |
TACGAGTCCTTCTGGCCCAT |
22 |
|
Beta tubulin |
Bt2a
(Fw) |
GGTAACCAAATCGGTGCTGCTTTC |
23 |
|
(BT2) |
Bt2b
(Rw) |
ACCCTCAGTGTAGTGACCCTTGGC |
23 |
|
Calmodulin |
cmd5
(Fw) |
CCGAGTACAAGGAGGCCTTC |
18 |
|
(CAL) |
cmd6
(Rw) |
CCGATAGAGGTCATAACGTGG |
18 |
3. Identification using phylogenetic analysis of DNA sequences
The sequences of each gene resulted from Sanger's sequencing were edited with BIOEDIT version 7.2.524 and blasted against the type materials and some reference strains in GenBank with a megablast in BLAST® blastn (https:// blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_SPEC=GeoBlast&PAGE_TYPE=BlastSearch). The datasets, including the sequences of the tested strain and those resulting from the megablast, were prepared by BIOEDIT version 7.2.5 and aligned with the algorithms, i.e., ClustalW in BIOEDIT version 7.2.5 or Multiple Alignment with Fast Fourier Transform (MAFFT) or MUltiple Sequence Comparison by Log-Expectation (MUSCLE)25,26 in EMBI-EBI web server (https://www.ebi.ac.uk/jdispatcher/). Phylogenetic analyses for identification had been conducted with MEGA X27. The best model and appropriate phylogenetic algorithms were inferred to reconstruct a phylogenetic tree and to estimate percentage similarity.
4. Antifungal susceptibility test
An AST kit, Thermo ScientificTM SensititreTM YeastOneTM YO10 AST Plate (Thermo Scientific Inc., UK), was used to test 12 Aspergillus strains and a reference strain, A. fumigatus ATCC-MYA-3626. The inoculum was prepared with a CLSI M38-A2 protocol. The conidial inoculum concentration was approximately 0.4 × 104 to 5 × 104 CFU/ml. Each strain was tested against 9 antifungal drugs, i.e., anidulafungin (AND), caspofungin (CAS), micafungin (MF), amphotericin B (AB), flucytosine (FC), fluconazole (FZ), itraconazole (IZ), posaconazole (PZ), and voriconazole (VZ). The AST plates were kept at 28-30℃, and the minimal effective concentration (MEC) values were observed after incubation for 24 h by microscopy examination. The minimal inhibitory concentration (MIC) values were observed after 48 h of incubation by a Vizion system, Thermo ScientificTM SensititreTM VizionTM Digital MIC Viewing System (Thermo Scientific Inc., UK).
1. Morphological identification and growth characters
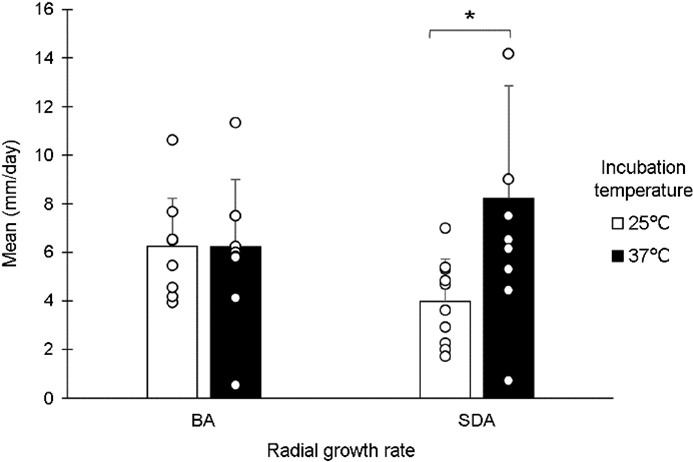
Table 2 presents 12 morphologically identified Aspergillus strains, together with their associated information. All strains grew at 25℃ and 37℃ and were significantly better on SDA at 37℃ but worse at 25℃ (p < 0.05) (Fig. 1). An FG-090 strain demonstrated the fastest growth on BA but an FG-091 strain exhibited the slowest on SDA at 25℃. FG-026, FG-054, and FG-069 strains exhibited the fastest growth on SDA but an FG-088 strain demonstrated the slowest on BA at 37℃ (Fig. 2).
|
No. |
Patients' information |
Diag.† |
Treat‡ |
Specimen site§ |
Patho. exam.∥ |
PCR¶ |
Fungal
cultivation |
|||
|
Sex |
Age |
Under. |
Morpho. Ident.** |
Strain No. |
||||||
|
1 |
F |
60 |
UD |
FK |
PKP |
Lt |
+ |
+ |
A. fumigatus |
FG-026 |
|
2 |
M |
52 |
OT |
FK |
EV |
Lt |
ND |
ND |
A. flavus |
FG-033 |
|
3 |
M |
40 |
UD |
FK |
EV |
Rt |
+ |
ND |
Aspergillus sp. |
FG-050 |
|
4 |
M |
39 |
OT |
FK |
EV |
Lt |
+ |
ND |
A. fumigatus |
FG-054 |
|
5 |
M |
63 |
UD |
FK |
PKP |
Lt |
+ |
ND |
A. flavus |
FG-060 |
|
6 |
M |
67 |
UD |
FK |
PKP |
Rt |
ND |
+ |
Aspergillus sp. |
FG-069 |
|
7 |
M |
44 |
DM |
FK |
EV |
Rt |
+ |
ND |
A. flavus |
FG-074 |
|
8 |
F |
64 |
UD |
FK |
PKP |
Rt |
ND |
ND |
A. flavus |
FG-076 |
|
9 |
M |
40 |
OT |
ED |
EV |
Rt |
ND |
ND |
Aspergillus sp. |
FG-088 |
|
10 |
M |
74 |
UD |
FK |
EV |
Lt |
ND |
+ |
A. flavus |
FG-090 |
|
11 |
M |
43 |
UD |
FK |
Med. |
Lt |
ND |
+ |
A. terreus |
FG-091 |
|
12 |
F |
87 |
UD |
FK |
PKP |
Lt |
+ |
+ |
A. terreus |
FG-093 |
|
*Underlying
conditions: UD=undetermined, OT=ocular trauma, DM=Diabetes Mellitus †Diagnosis:
FK=fungal keratitis, ED=Endophthalmitis ‡Treatment:
PKP=Penetrating keratoplasty, EV=Evisceration, Med.=treatment with antifungal
drugs but lost the follow up §Specimen
site: Lt=left eye, Rt=right eye ∥Histopathological examination, ¶PCR of
fungal DNA in specimens: + = positive for fungal elements/fungal DNA, ND =
not determine **Morphological identification |
||||||||||
2. DNA barcoding in 12 Aspergillus strains
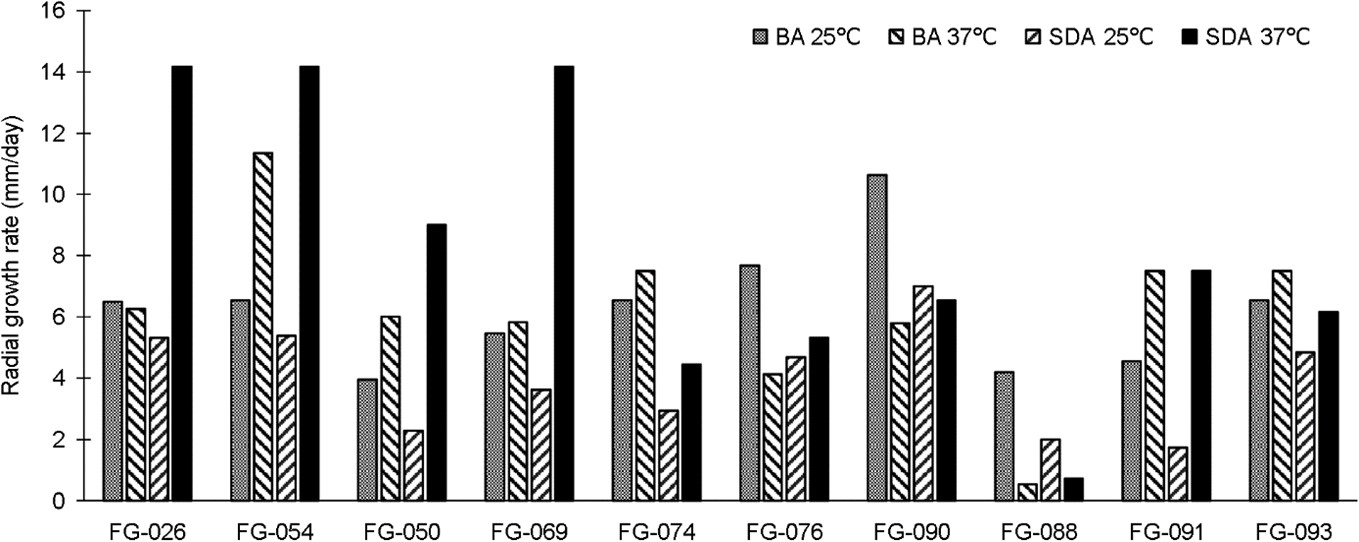
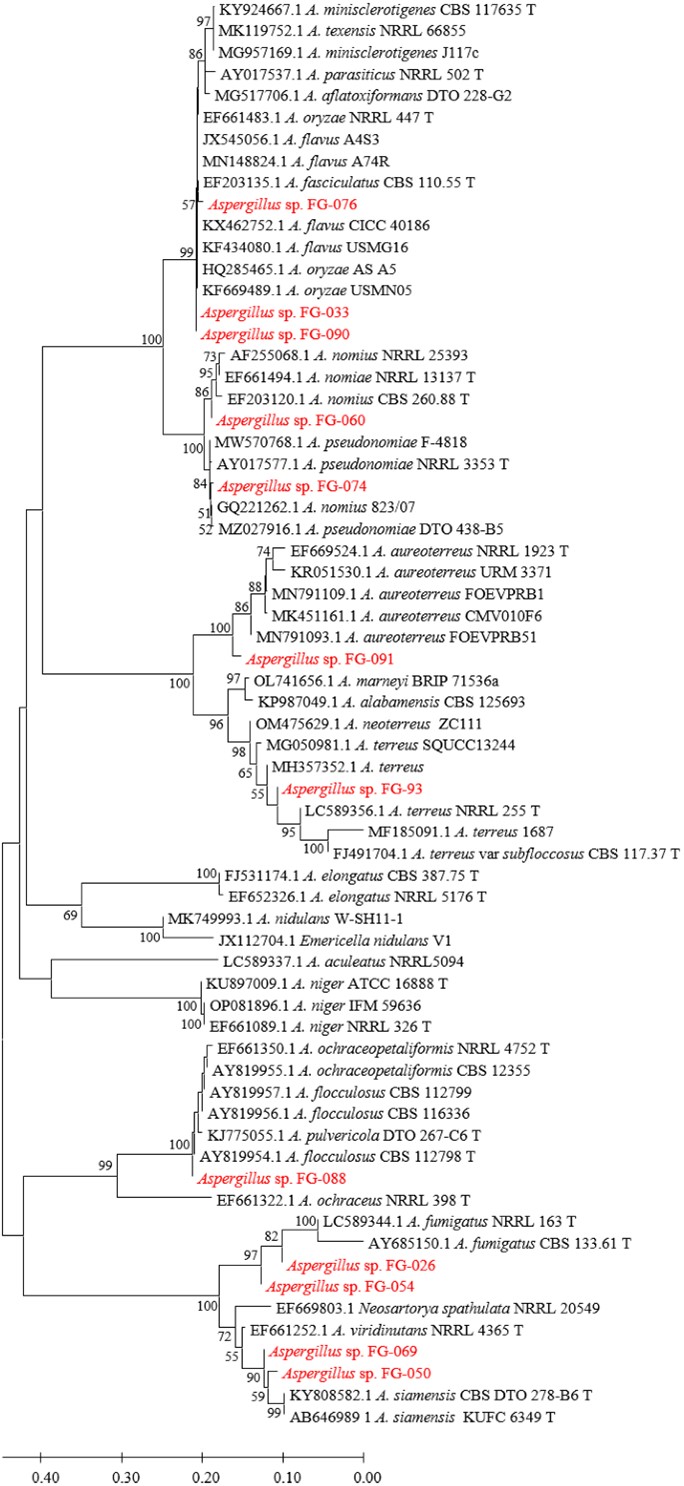
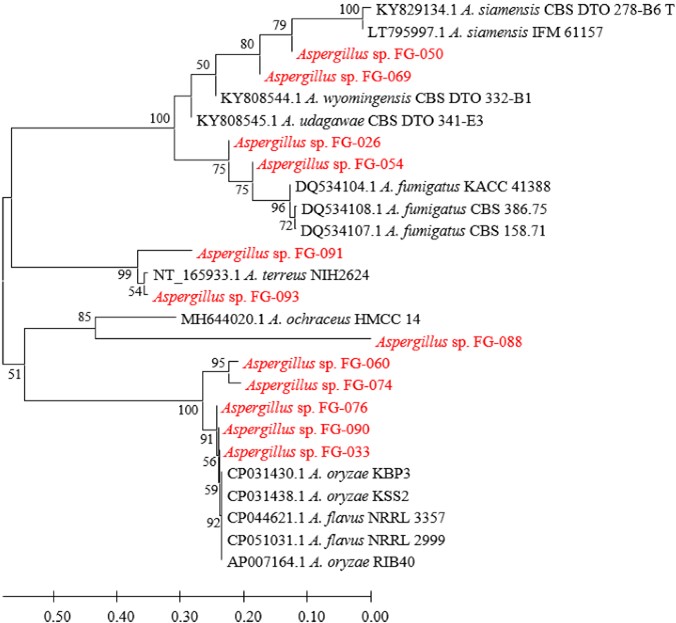
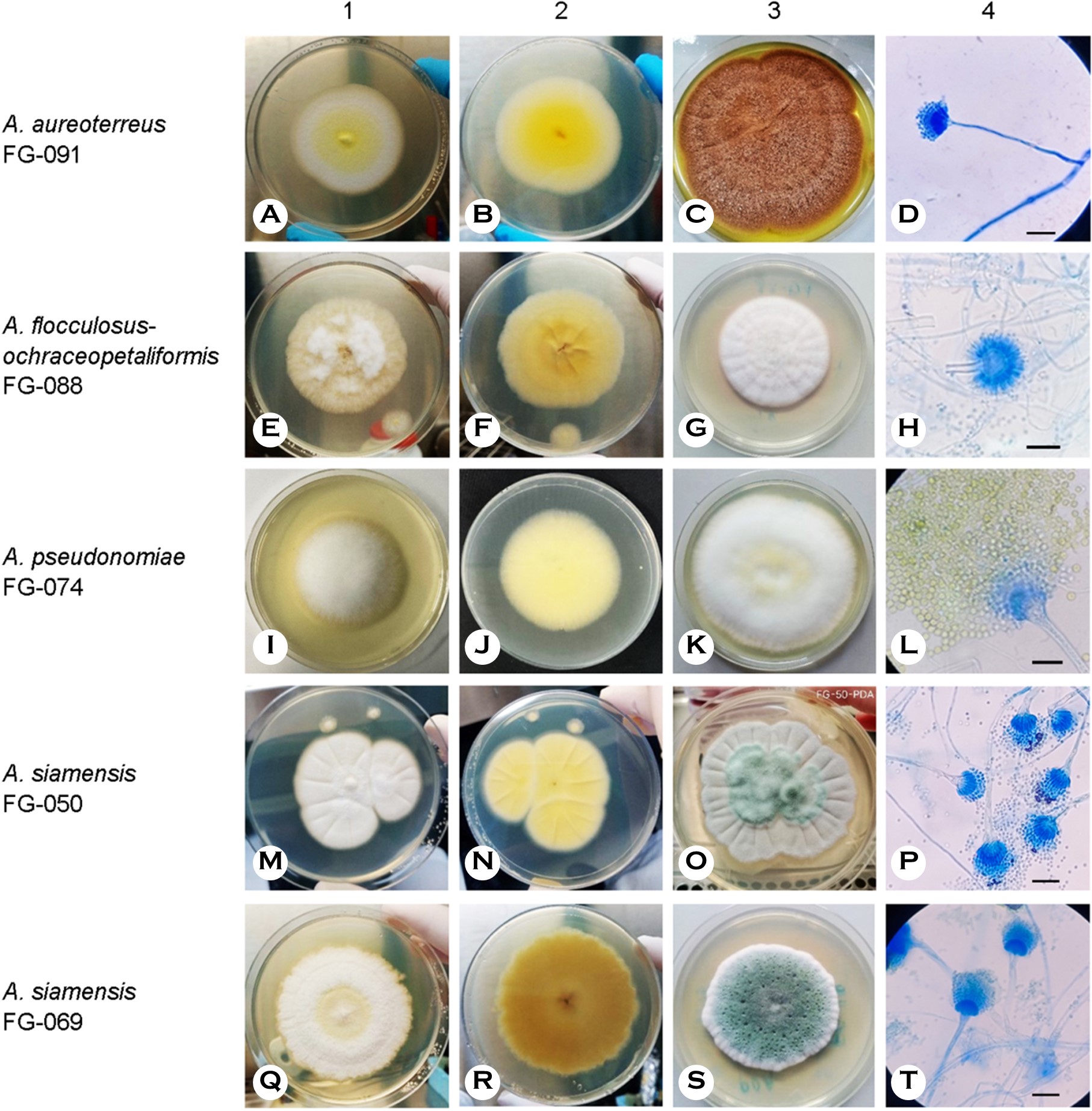
Figs. 3, 4, 5, and 6 show the phylogenetic analyses with ITS, BT2, CAL, and ACT, respectively. ITS, BT2 and CAL, and ACT identified 5, 6, and 3 species and 3, 2, and 1 species com- plex, respectively. Table 3 concludes all identification results.The multi-locus sequence typing (MLST) identification in- dicated A. aureoterreus, A. pseudonomiae, A. siamensis, and A. flocculosus-ochraceopetaliformis as the four new pathogens causing eye infection in human, and Fig. 7 illustrates their morphological characters.

|
Strain |
DNA Identification: the
highest similarity (%) comparing to the type or reference materials |
|||||||
|
ITS |
Species |
BT2 |
Species |
CAL |
Species |
ACT* |
Species |
|
|
FG-033 |
100% |
A. flavus cpx.† |
100% |
A. flavus cpx.‡ |
97.46~ |
A. flavus cpx.§ |
99.71% 99.71% |
A. flavus A. oryzae |
|
FG-076 |
100% |
A. flavus cpx.† |
99.18~ |
A. flavus cpx.‡ |
97.70~ |
A. flavus cpx.‡ |
99.71% 99.71% |
A. flavus A. oryzae |
|
FG-090 |
100% |
A. flavus cpx.† |
100% |
A. flavus cpx.‡ |
98.65~ |
A. flavus cpx.‡ |
99.71% 99.71% |
A. flavus A. oryzae |
|
FG-026 |
100% |
A. fumigatus |
100% |
A. fumigatus |
99.80% |
A. fumigatus |
99.73% |
A. fumigatus |
|
FG-054 |
100% |
A. fumigatus |
100% |
A. fumigatus |
100% |
A. fumigatus |
100% |
A. fumigatus |
|
FG-088 |
100% 100% |
A. flocculosus A. ochraceo-petaliformis |
100% 100% |
A. flocculosus A. ochraceo-petaliformis |
99.67~ 99.83% |
A. flocculosus A. ochraceo-petaliformis |
ND |
ND |
|
FG-060 |
100% |
A. nomiae |
99.60% |
A. nomiae |
99.83% |
A. nomiae |
ND |
ND |
|
FG-074 |
99.82% |
A. pseudonomiae |
99.79% |
A. pseudonomiae |
99.60% |
A. pseudonomiae |
ND |
ND |
|
FG-050 |
100% |
A. siamensis |
99.97% |
A. siamensis |
97.67% |
A. siamensis |
95.49% |
A. siamensis |
|
FG-069 |
100% |
A. siamensis |
99.98% |
A. siamensis |
97.67% |
A. siamensis |
95.83% |
A. siamensis |
|
FG-091 |
100% |
A. aureoterreus |
98.93% |
A. aureoterreus |
98.10% |
A. aureoterreus |
ND |
ND |
|
FG-093 |
100% |
A. terreus cpx.∥ |
100% |
A. terreus |
99.13% |
A. terreus |
98.95% |
A. terreus |
|
*limited number of type or reference
materials in a public database †including A. aflatoxiformans, A.
fasciculatus, A. flavus, A. minisclerotigenes and A. oryzae ‡including A. fasciculatus, A. flavus and
A. oryzae §including A. fasciculatus, A. flavus,
A. minisclerotigenes and A. oryzae ∥including A. terreus, A. neoterreus,
A. elongatus and A. montoensis |
||||||||
3. Antifungal susceptibility test
A. fumigatus ATCC-MYA 3626 revealed MIC values of all tested drugs with the AST kit in those ranges recommended by CLSI M61. The MICs of echinocandins and FZ of all tested strains were higher than the test set. However, their other MIC values exhibited differently in AB, FC, and other azoles. Table 4 presents all MEC and MIC values of the 12 strains.
|
Strain |
Species |
MEC (μg/ml) |
|
MIC (μg/ml) |
||||||||
|
AND |
AND |
CAS |
MF |
AB |
FC |
FZ |
IZ |
PZ |
VZ |
|||
|
FG-033 |
A. flavus cpx. |
≤0.015 |
|
>8 |
>8 |
>8 |
2 |
>64 |
>256 |
0.50 |
0.50 |
1 |
|
FG-076 |
A. flavus cpx. |
≤0.015 |
|
>8 |
>8 |
>8 |
4 |
>64 |
>256 |
0.25 |
0.25 |
0.50 |
|
FG-090 |
A. flavus cpx. |
≤0.015 |
|
>8 |
>8 |
>8 |
4 |
>64 |
>256 |
0.25 |
0.50 |
1 |
|
FG-026 |
A. fumigatus |
≤0.015 |
|
>8 |
>8 |
>8 |
2* |
>64 |
>256 |
0.25 |
0.12 |
0.50 |
|
FG-054 |
A. fumigatus |
≤0.015 |
|
>8 |
>8 |
>8 |
1 |
>64 |
>256 |
0.25 |
0.12 |
0.25 |
|
FG-088 |
A. flocculosus- |
≤0.015 |
|
>8 |
>8 |
>8 |
8† |
16‡ |
>256 |
0.50 |
0.50 |
0.50 |
|
FG-060 |
A. nomiae |
≤0.015 |
|
>8 |
>8 |
>8 |
2 |
32 |
>256 |
0.50 |
0.50 |
2† |
|
FG-074 |
A. pseudonomiae |
≤0.015 |
|
>8 |
>8 |
>8 |
2 |
>64 |
>256 |
0.25 |
0.25 |
0.25 |
|
FG-050 |
A. siamensis |
≤0.015 |
|
>8 |
>8 |
>8 |
2† |
>64 |
>256 |
0.50 |
0.50 |
2† |
|
FG-069 |
A. siamensis |
≤0.015 |
|
>8 |
>8 |
>8 |
4† |
>64 |
>256 |
1 |
0.50 |
4† |
|
FG-091 |
A. aureoterreus |
≤0.015 |
|
>8 |
>8 |
>8 |
4 |
2‡ |
>256 |
0.25 |
0.12 |
0.25 |
|
FG-093 |
A. terreus |
≤0.015 |
|
>8 |
>8 |
>8 |
2 |
>64 |
>256 |
0.12 |
0.12 |
0.12 |
|
ATCC- MYA-3626 |
A. fumigatus |
≤0.015 |
|
>8 |
>8 |
>8 |
2* |
>64 |
>256 |
0.50 |
0.25 |
0.50 |
|
MEC=minimal effective
concentration, MIC=minimal inhibitory concentration, AND=anidulafungin, CAS=caspofungin,
MF=micafungin, AB= amphotericin B, FC=flucytosine, FZ=fluconazole,
IZ=itraconazole, PZ=posaconazole, VZ=voriconazole * = resistant, † = likely resistant (no breakpoint
proposed), ‡ = likely
sensitive (no breakpoint proposed) |
||||||||||||
In Thailand, infectious keratitis was annually estimated to occur in 1 per 1,000 people, and approximately 15% of those were FK. Thailand would have an estimated 70,000 patients with infectious keratitis, with approximately 10,500 cases of FK28. Aspergillus species is one of the most prevalent pathogens of FK9,10. A total of 73 FK cases of mold infection were reported in the Srinagarind Hospital, Khon Kaen, Thailand, from January 1 to December 31, 2022, in which 12 (16.44%) cases were caused by Aspergillus spp. The incidence is <21.5% and 17.5% of those from India and Ghana12, respectively. However, the real number of burden cases of Aspergillus eye infection is possibly higher than the estimation based on the number of hospitals all around Thailand.
The systematic classification of most fungal species relies on the genealogical species concept with MLST. Genes, such as ITS, and partial sequences of BT2, CAL, and ACT, have been the barcoding markers to identify Aspergillus species by DNA sequences29-31. ITS has been the primary barcoding marker because of the most available sequence in the public databases. However, it demonstrates less specificity for some Aspergillus species such as in A. flavus complex and A. terreus complex. Therefore, the "secondary barcoding markers" viz. BT2, CAL, and ACT have been employed. These markers cannot completely identify the A. flavus complex, but they successfully determined A. terreus. Presently, these secondary barcoding markers, especially, of some type or reference materials have been unavailable in the public database. All potential barcoding marker sequences of the type and/or reference materials should be generated and submitted into the public database to support the identification of the DNA sequences.
The present study identified 6 Aspergillus species and 2 species complex causing eye infections viz. A. flavus complex, A. fumigatus, A. terreus, A. aureoterreus, A. nomiae, A. pseudonomiae, A. siamensis, and A. flocculosus-ochraceo- petaliformis by PCR, Sanger sequencing, and phylogenetic methods. A. flavus, A. fumigatus, and A. terreus have usually been reported as common species12-15. A. nomiae, a neglected species that causes FK, has been reported16. No case of eye infection caused by A. aureoterreus, A. pseudonomiae, A. siamensis, and A. flocculosus-ochraceopetaliformis has been previously published. Therefore, this investigation revealed the four new pathogens of eye infection.
Samson, S.W. Peterson, Frisvad & Varga raised A. aureoterreus from A. terreus var aureus by a yellow pigment producing and a distant genotype from A. terreus32. ITS, BT2, and CAL are barcoding markers for species identification. A. aureoterreus can be found in soil or as a contaminant in wheat flour32 and spice34. The species was mentioned as one isolation from a patient but with no pathological in- formation15. Its morphology and growth character are closely similar to those of A. terreus. Concurrently, A. terreus and A. aureoterreus might be misidentified when DNA identification was not available to determine the fungal isolates from many A. terreus infection cases. A. aureoterreus strain FG-091 (Fig. 7A-D) has presently been the first isolated from a corneal scraping of a patient with keratitis. The fungal DNA could be detected by PCR although the specimens did not reveal fungal elements by pathological examination. The strain demonstrated better growth at 37℃, the same as A. terreus FG-093, but with a colony producing a yellow pigment (Fig. 7C). The patient was treated with antifungal drugs but the outcome remains unknown.
Varga, Samson & Frisvad isolated A. pseudonomiae from the soil, insect33, and house dust35. ITS and BT2 can be utilized as barcoding markers for this species. The species was proposed because of genetic differences from A. nomiae. Similar to A. nomiae, it can be morphologically misidentified as A. flavus. Manikandan et al.16 isolated Aspergillus sp. strain 823/07 from a keratitis case and identified the fungus as A. nomiae by BLASTN search. However, strain 823/07 was later identified as A. pseudonomiae by phylogenetic analysis of the concatenated sequence of ITS, BT2, and CAL35. Our inves- tigation confirmed the strain 823/07 to be A. pseudonomiae by phylogenetic analyses with only a single gene, exhibiting the highest similarities to the type material, i.e., 99.80% of ITS, 99.62% of BT2 and 99.46% of CAL. Consequently, A. pseudonomiae strain FG-074 (Fig. 7I-L) is officially the first isolation from the patient with keratitis but unofficially, the second of the reported cases. The strain FG-074 was isolated from the right eye of a patient with diabetes mellitus. Histo- pathological examination confirmed positive fungal elements. The strain exhibited good growth at 37℃ and morphological characters similar to those of the A. nomiae FG-060 strain and the A. flavus group. The patient demonstrated a severe prognosis which may be associated with the underlying condition, with evisceration as the outcome.
Manoch, Eamvijarn & Yaguchi36 first isolated A. siamensis from coastal forest soil in Thailand and classified them in Aspergillus section Fumigati, with no other habitat being mentioned since then. ITS, BT2, CAL, and ACT are barcoding markers for species identification. The species is closely asso- ciated with A. udagawae and A. viridinutans which have previously been reported their pathogenicity37-39. A. siamensis first appeared as a pathogen in Japan, causing a fatal dis- seminated infection in an 18-year-old male patient. The patient received bone marrow transplantation after having chronic granulomatous disease for a long time40. Two strains of A. siamensis, including FG-050 (Fig. 7M-P) and FG-069 (Fig. 7Q-T), were isolated from two patients with FK. The cases have been the second and the third of the pathogenicity finding but the first of eye infection. The FG-050 strain was isolated from a corneal scraping specimen of a right eye as well as fungal positive by both histopathological staining and PCR for fungal DNA in specimens. The patient was not successfully treated with the medication; hence, evisceration was required. Another strain, the FG-069, was isolated from a cornea specimen of a right eye infection. No histopathological staining was conducted but PCR for fungal DNA was positive. The patient was treated with penetrating keratoplasty. Both FG-050 and FG-069 strains grew better on SDA, especially at 37℃, similar to the type strain36 and the important pathogens such as A. fumigatus and A. flavus. The ability to grow well at 37℃ may be a criterion for the pathogenicity prediction of Aspergillus species.
Frisvad & Samson classified A. flocculosus in the Aspergillus section Circumdati or A. ochraceus group. It could be isolated from the saltern, soil, and substratum in tropical and temperate areas41. The species internally infected grape35 but with no reported human infection. Conversely, Batista & da Silva Maia isolated A. ochraceopetaliformis from a scalp lesion in a Brazilian man42 (a type strain CBS 123.55) (https:// www.mycobank.org) and reported of causing nail infection in a German woman (strain CBS 123362)43. A. flocculosus and A. ochraceopetaliformis were noted to have been genetically very similar to each other41,44. Defining the right taxa of both species is beyond our investigation. Therefore, this study only designated the strain FG-088 (Fig. 7E-H) in a complex species as A. flocculosus-ochraceopetaliformis. The strain FG-088 grew slower and did not grow well at 37℃, unlike other Aspergillus pathogens. It was isolated from a cornea specimen of the right eye in a patient with eye injury caused by wood puncture and developed endophthalmitis. Histopathological test and PCR for direct detection was not investigated. The patient was treated with enucleation because of a very poor prognosis which may be from severe injury and infection.
The sources of the five strains in the environment around each patient remain unclear. The predisposing factors, such as accidental evidence causing eye injury, trauma, or underlying condition, have to be considered. Notably, the virulence of Aspergillus keratitis demonstrated a lower successful treatment rate with slow healing, and a failure of primary treatment was at a higher rate than keratitis by Fusarium and other filamentous fungi12. The five pathogens exhibited their virulence concordant to the previous investigation.
The MIC-breakpoints of some species and some antifungal drugs proposed by CLSI45 and EUCAST46 revealed that only A. fumigatus FG-026 was resistant to AB (>1 μg/ml). Con- versely, all of them were likely sensitive to IZ (≤1 μg/ml). Aspergillus section Flavi, A. flavus and A. nomiae, seemed to be resistant to PZ (>0.25 μg/ml). Echinocandins with a low MEC (0.015 μg/ml) could affect the growth of all Aspergillus strains, but they could not inhibit the growth of all strains at the highest concentration. The similar result was nearly the same as FZ which could not inhibit the growth of all tested strains. Notably, the four new pathogens likely demon- strated distinguished susceptibility profiles. A. flocculosus-ochraceopetaliformis strain FG-088 was resistant against AB with the highest MIC (8 μg/ml), but sensitive to FC. Further, the strain was resistant to FZ but sensitive to IZ, as A. ochraceopetaliformis mentioned in a previous report43. Both A. siamensis strains were likely resistant to VZ (>2 μg /ml) with the highest MIC of 4 μg/ml. The phenomenon may be inherited because it is similar to the resistant MICs of other members classified in the sections, Circumdati and Fumigati, that was previously investigated47. A. aureoterreus strain FG-091 was susceptible to FC with the lowest MIC. FC has barely been used alone to treat Aspergillus eye infection but a combination with topical AB demonstrated the synergistic effect in FK with yeast infection48. However, the MIC profiles of each strain should have been further assessed, especially associations with the genotypic characters.
In conclusion, the DNA determination with a sequence of a single gene as a barcoding tool for identifying Aspergillus species exhibited more satisfactory results than the morphological identification. The method could not determine some closely related members of the A. flavus complex. However, because of better precision and accuracy, it remains strongly recommended in routine identification. Moreover, PCR and sequencing facilities have been presently simplified, with a shorter turnaround time. Consequently, the DNA method would be beneficial to appropriately treat a patient, support a high rate of successful treatment, and finally, reliably report the case.
References
1. Perlin DS, Wiederhold NP. Culture-independent molecular methods for detection of antifungal resistance mech- anisms and fungal identification. J Infect Dis 2017;216: S458-S465
Google Scholar
2. Wickes BL, Wiederhold NP. Molecular diagnostics in medical mycology. Nat Commun 2018;9:5135
Google Scholar
3. Manikandan P, Abdel-Hadi A, Randhir Babu Singh Y, Revathi R, Anita R, Banawas S, et al. Fungal Keratitis: epidemiology, rapid detection, and antifungal suscepti- bilities of Fusarium and Aspergillus isolates from corneal scrapings. Biomed Res Int 2019:6395840
Google Scholar
4. Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel) 2017;3:57
Google Scholar
5. Thomas PA, Kaliamurthy J. Mycotic keratitis: epidemiology, diagnosis and management. Clin Microbiol Infect 2013; 19:210-220
Google Scholar
6. Srinivasan R, Kanungo R, Goyal JL. Spectrum of oculo- mycosis in south India. Acta Ophthalmol (Copenh) 1991; 69:744-749
Google Scholar
7. Lakhundi S, Siddiqui R, Khan NA. Pathogenesis of micro- bial keratitis. Microb Pathog 2017;104:97-109
Google Scholar
8. Ghosh AK, Gupta A, Rudramurthy SM, Paul S, Hallur VK, Chakrabarti A. Fungal keratitis in north India: spectrum of agents, risk factors and treatment. Mycopathologia 2016;181:843-850
Google Scholar
9. Thomas PA, Leck AK, Myatt M. Characteristic clinical features as an aid to the diagnosis of suppurative keratitis caused by filamentous fungi. Br J Ophthalmol 2005;89: 1554-1558
Google Scholar
10. Kredics L, Narendran V, Shobana CS, Vágvölgyi C, Manikandan P, Indo-Hungarian Fungal Keratitis Working Group. Filamentous fungal infections of the cornea: a global overview of epidemiology and drug sensitivity. Mycoses 2015;58:243-260
Google Scholar
11. Lalitha P, Prajna NV, Kabra A, Mahadevan K, Srinivasan M. Risk factors for treatment outcome in fungal keratitis. Ophthalmology 2006;113:526-530
Google Scholar
12. Leck AK, Thomas PA, Hagan M, Kaliamurthy J, Ackuaku E, John M, et al. Aetiology of suppurative corneal ulcers in Ghana and south India, and epidemiology of fungal keratitis. Br J Ophthalmol 2002;86:1211-1215
Google Scholar
13. Kunimoto DY, Sharma S, Garg P, Gopinathan U, Miller D, Rao GN. Corneal ulceration in the elderly in Hyderabad, south India. Br J Ophthalmol 2000;84:54-59
Google Scholar
14. Manikandan P, Dóczi I, Kocsubé S, Varga J, Németh TM, Antal Z, et al. Aspergillus species in human kerato- mycosis. In: Varga J, Samson RA, editors. Aspergillus in the genomic era. Wageningen: Academic Publishers, 2008:293-328
Google Scholar
15. Gautier M, Normand AC, Ranque S. Previously unknown species of Aspergillus. Clin Microbiol Infect 2016;22: 662-669
Google Scholar
16. Manikandan P, Varga J, Kocsubé S, Samson RA, Anita R, Revathi R, et al. Mycotic keratitis due to Aspergillus nomius. J Clin Microbiol 2009;47:3382-3385
Google Scholar
17. Shigeyasu C, Yamada M, Nakamura N, Mizuno Y, Sato T, Yaguchi T. Keratomycosis caused by Aspergillus viridinutans: an Aspergillus fumigatus-resembling mold presenting distinct clinical and antifungal susceptibility patterns. Med Mycol 2012;50:525-528
Google Scholar
18. Karimizadeh Esfahani M, Eslampoor A, Dolatabadi S, Najafzadeh MJ, Houbraken J. First case of fungal keratitis due to Aspergillus minisclerotigenes in Iran. Curr Med Mycol 2019;5:45-48
Google Scholar
19. de Hoog GS, Gerrits van den Ende AH. Molecular diag- nostics of clinical strains of filamentous Basidiomycetes. Mycoses 1998;41:183-189
Google Scholar
20. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A guide to methods and applications. New York: Academic Press, 1989:315-322
Google Scholar
21. Masclaux F, Guého E, de Hoog GS, Christen R. Phylo- genetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. J Med Vet Mycol 1995;33:327-338
Google Scholar
22. Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999;91:553-556
Google Scholar
23. Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 1995;61:1323-1330
Google Scholar
24. Hall TA. BioEdit: a user-friendly biological sequence align- ment editor and analysis program for Windows 95/98/ NT. In: Nucleic acids symposium series no. 41. Oxford University Press, 1999:95-98
Google Scholar
25. Katoh K, Standley DM. MAFFT multiple sequence align- ment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772-780
Google Scholar
26. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792-1797
Google Scholar
27. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-1549
Google Scholar
28. Chayakulkeeree M, Denning DW. Serious fungal infec- tions in Thailand. Eur J Clin Microbiol Infect Dis 2017; 36:931-935
Google Scholar
29. Kidd S, Halliday C, Alexiou H, Ellis D. Descriptions of medical fungi, 3rd ed. Adelaide: Newstyle Printing, 2016: v-vi
Google Scholar
30. Hinrikson HP, Hurst SF, De Aguirre L, Morrison CJ. Molecular methods for the identification of Aspergillus species. Med Mycol 2005;43:S129-S137
Google Scholar
31. Tam EW, Chen JH, Lau EC, Ngan AH, Fung KS, Lee KC, et al. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: characterization by internal transcribed spacer, β-tubulin, and calmodulin gene se- quencing, metabolic fingerprinting, and matrix-assisted laser desorption ionization-time of flight mass spectro- metry. J Clin Microbiol 2014;52:1153-1160
Google Scholar
32. Samson RA, Peterson SW, Frisvad JC, Varga J. New species in Aspergillus section Terrei. Stud Mycol 2011;69:39-55
Google Scholar
33. Varga J, Frisvad JC, Samson RA. Two new aflatoxin pro- ducing species, and an overview of Aspergillus section Flavi. Stud Mycol 2011;69:57-80
Google Scholar
34. Mahata PK, Dass RS, Pan A, Muthusamy B. Substantive Morphological descriptions, phylogenetic analysis and single nucleotide polymorphisms of Aspergillus species from Foeniculum vulgare. Front Microbiol 2022;13: 832320
Google Scholar
35. Zhou YB, Li DM, Houbraken J, Sun TT, de Hoog GS. Fatal rhinofacial mycosis due to Aspergillus nomiae: case report and review of published literature. Front Microbiol 2020; 11:595375
Google Scholar
36. Eamvijarn A, Manoch L, Chamswarng C, Piasai O, Visarathanonth N, Luangsa-ard JJ, et al. Aspergillus siamensis sp. nov. from soil in Thailand. Mycoscience 2013;54:401-405
Google Scholar
37. Vinh DC, Shea YR, Sugui JA, Parrilla-Castellar ER, Freeman AF, Campbell JW, et al. Invasive aspergillosis due to Neosartorya udagawae. Clin Infect Dis 2009;49:102-111
Google Scholar
38. Posteraro B, Mattei R, Trivella F, Maffei A, Torre A, De Carolis E, et al. Uncommon Neosartorya udagawae fungus as a causative agent of severe corneal infection. J Clin Microbiol 2011;49:2357-2360
Google Scholar
39. Coelho D, Silva S, Vale-Silva L, Gomes H, Pinto E, Sarmento A, et al. Aspergillus viridinutans: an agent of adult chronic invasive aspergillosis. Med Mycol 2011; 49:755-759
Google Scholar
40. Maemura R, Wakamatsu M, Sakaguchi H, Yoshida N, Karakawa S, Kobayashi M, et al. Disseminated Aspergillus siamensis infection following haploidentical bone marrow transplantation for chronic granulomatous disease. Rinsho Ketsueki 2020;61:327-333
Google Scholar
41. Frisvad JC, Frank JM, Houbraken JAMP, Kuijpers AF, Samson RA. New ochratoxin A producing species of Aspergillus section Circumdati. Stud Mycol 2004;50:23-43
Google Scholar
42. Batista AC, Maia HS. Alguns Aspergillus contaminantes de culturas. Anais Soc Biol Pernamb 1957;15:181-237
Google Scholar
43. Brasch J, Varga J, Jensen JM, Egberts F, Tintelnot K. Nail infection by Aspergillus ochraceopetaliformis. Med Mycol 2009;47:658-662
Google Scholar
44. Peterson SW. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 2008; 100:205-226
Google Scholar
45. Clinical and Laboratory Standard Institute (CLSI). Perform- ance Standards for Antifungal Susceptibility Testing of Filamentous Fungi. 3rd ed. CLSI supplement M38M51S. Clinical and Laboratory Standards Institute, USA, 2022
46. The European committee on antimicrobial susceptibility testing. Overview of antifungal ECOFFs and clinical break- points for yeasts, moulds and dermatophytes using the EUCAST E.Def 7.4, E.Def 9.4 and E.Def 11.0 procedures. Version 4.0, 2023
47. Wang HC, Hsieh MI, Choi PC, Wu WL, Wu CJ; TSARM Hospitals. Species distribution and antifungal susceptibility of clinical Aspergillus isolates: A multicentre study in Taiwan, 2016-2020. Mycoses 2023;66:711-722
Google Scholar
48. Suman S, Kumar A, Saxena I, Kumar M. Fungal keratitis: Recent advances in diagnosis and treatment. In: Rodriguez-Garcia A, Hernandez-Camarena JC, editors. Infectious Eye Diseases. London: IntechOpen Limited, 2021
Google Scholar
Congratulatory MessageClick here!